| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0014s15660 (EnsemblPlants link) | unknown (PX0011_B08) | Populus trichocarpa | ABK92452.1 (NCBI query) | A9P7U9(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0014s15660; this unknown (PX0011_B08) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0009s16530 (EnsemblPlants link) | WS0116_O03 | Populus trichocarpa | ABK94377.1 (NCBI query) | A9PDC4(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0009s16530; this WS0116_O03 transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
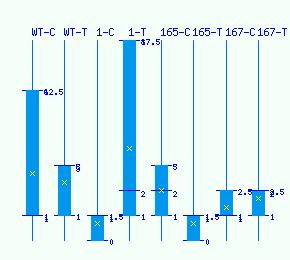
| GH17 | POPTR_0001s26210 (EnsemblPlants link) | unknown (WS0122_H18) | Populus trichocarpa | ABK95326.1 (NCBI query) | A9PG23(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Normalized RNA-seq library counts for POPTR_0001s26210
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0004s16120 (EnsemblPlants link) | unknown (WS0122_I17) | Populus trichocarpa | ABK95331.1 (NCBI query) | A9PG28(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0004s16120; this unknown (WS0122_I17) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0018s12960 (EnsemblPlants link) | unknown (WS0122_P13) (fragment) | Populus trichocarpa | ABK95381.1 (NCBI query) | A9PG78(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0018s12960; this unknown (WS0122_P13) (fragment) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0008s05600 (EnsemblPlants link) | unknown (WS0124_J02) | Populus trichocarpa | ABK95730.1 (NCBI query) | A9PH77(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0008s05600; this unknown (WS0124_J02) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH17 | POPTR_0009s11830 (EnsemblPlants link) | unknown (WS0125_F18) | Populus trichocarpa | ABK95837.1 (NCBI query) | A9PHI4(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0009s11830; this unknown (WS0125_F18) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||