| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table |
| GH19 | chitinase B | Populus trichocarpa | CAA42612.1 (NCBI query) | P29031(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | |
| No ORF identifier associated with this protein ID. | ||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH19 | POPTR_0009s14390 (EnsemblPlants link) | chitinase C (fragment) | Populus trichocarpa | CAA42614.1 (NCBI query) | P29032(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0009s14390; this chitinase C (fragment) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GH19 | POPTR_0010s15150 (EnsemblPlants link) | unknown (PX0011_C09) | Populus trichocarpa | ABK92465.1 (NCBI query) | A9P7W2(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
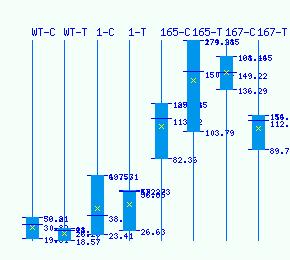
Normalized RNA-seq library counts for POPTR_0010s15150
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table |
| GH19 | unknown (WS0124_E20) | Populus trichocarpa | ABK95688.1 (NCBI query) | A9PH35(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | |
| No ORF identifier associated with this protein ID. | ||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||
| GH19 | POPTR_0014s14370 (EnsemblPlants link) | unknown (WS0125_J03) | Populus trichocarpa | ABK95866.1(NCBI query) | A9PHL3(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||
|
No RNA-seq library record found for POPTR_0014s14370; this unknown (WS0125_J03) transcript may have been excluded from differential analysis due to mapping non-uniquely to the P. trichocarpa reference (this prevents inflation of transcript counts). This can occur in families with very high (>99%) homology. | ||||||||