| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GT43 | POPTR_0006s13320 (EnsemblPlants link) | glycosyltransferase GT43A (GT43A ) | Populus trichocarpa | AEG25423.1 (NCBI query) | B9MT92(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
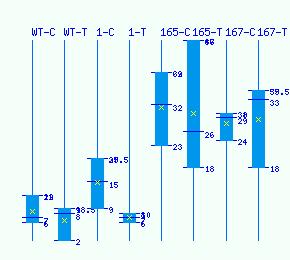
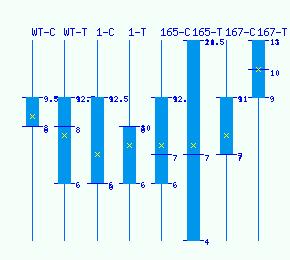
Normalized RNA-seq library counts for POPTR_0006s13320
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GT43 | POPTR_0016s08770 (EnsemblPlants link) | glycosyltransferase GT43B (GT43B ) | Populus trichocarpa | AEG25424.1 (NCBI query) | B9IJ10(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
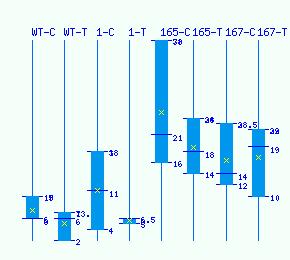
Normalized RNA-seq library counts for POPTR_0016s08770
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GT43 | POPTR_0007s10660 (EnsemblPlants link) | glycosyltransferase GT43C (GT43C) | Populus trichocarpa | AEG25425.1 (NCBI query) | B9HH87(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
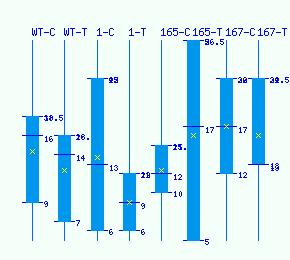
Normalized RNA-seq library counts for POPTR_0007s10660
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GT43 | POPTR_0005s18500 (EnsemblPlants link) | glycosyltransferase GT43D (GT43D) | Populus trichocarpa | AEG25426.1 (NCBI query) | F6M1G6(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
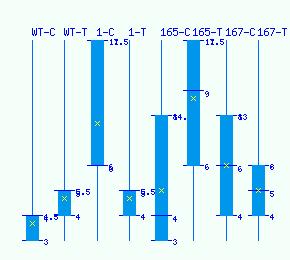
Normalized RNA-seq library counts for POPTR_0005s18500
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Gene | Protein name | Organism | Genbank ID | UniProt ID | Amino acid composition table | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GT43 | POPTR_0002s10790 (EnsemblPlants link) | glycosyltransferase GT43E (GT43E) | Populus trichocarpa | AEG25427.1 (NCBI query) | F6M1G7(UniProtKB link) | Amino Acid Composition(ExPASy link) Protein sequence(FASTA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Normalized RNA-seq library counts for POPTR_0002s10790
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||